Kocher Lab Online Resources¶
Specimen Database¶
The Kocher lab specimen database may be found at the Kocher Lab Specimen Database. By default, the login page will be displayed.
Access to the database requires:
- A Google account or a Princeton account (must be linked to Google)
- Permission for the account to access the database (ask Andrew)
Once granted access to the database, the homepage will be displayed.
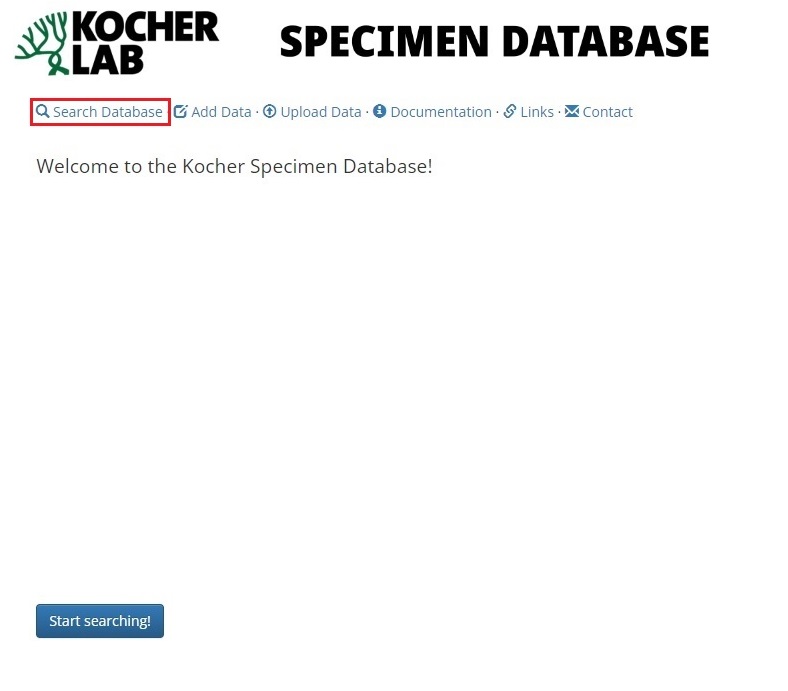
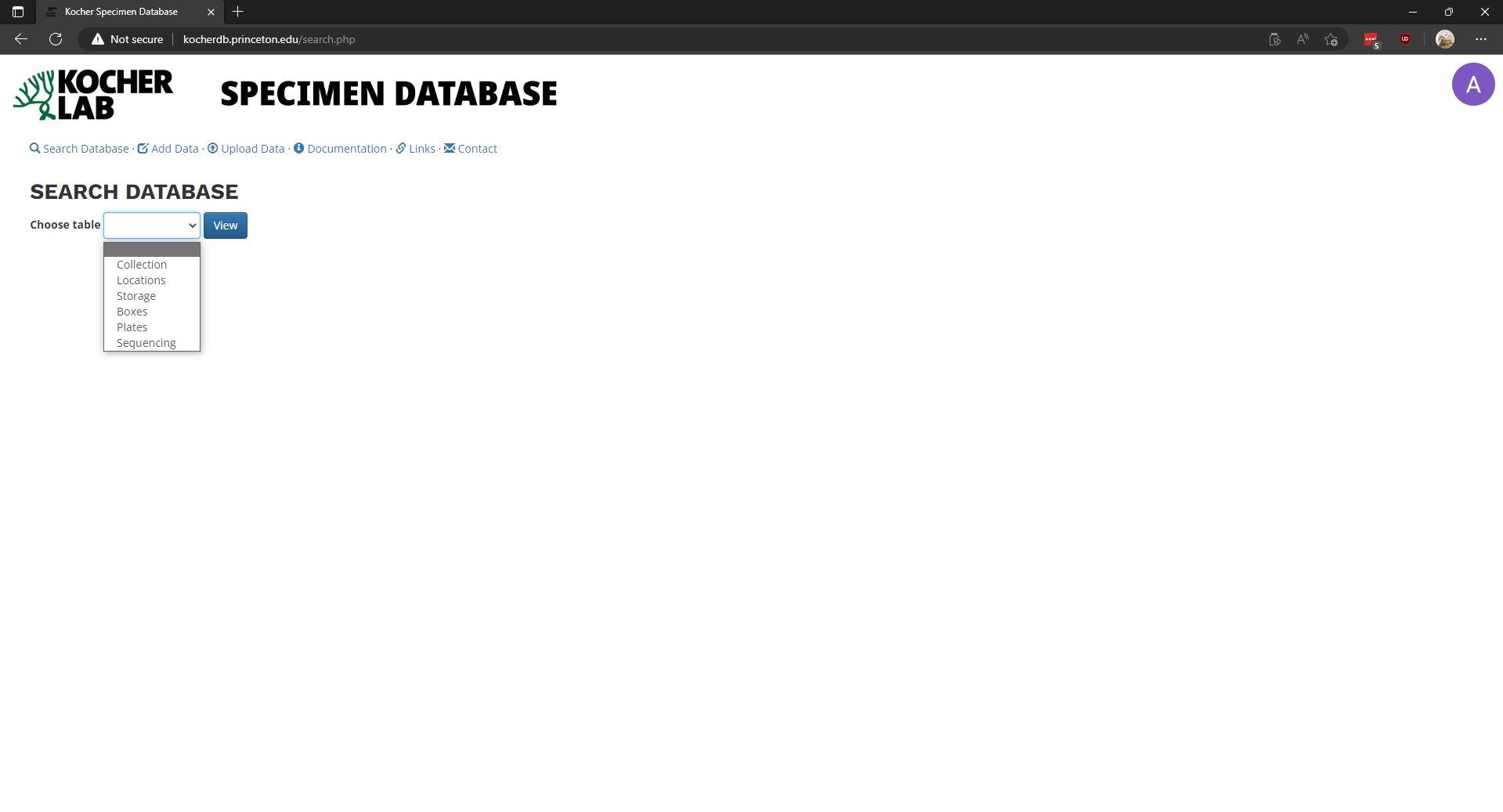
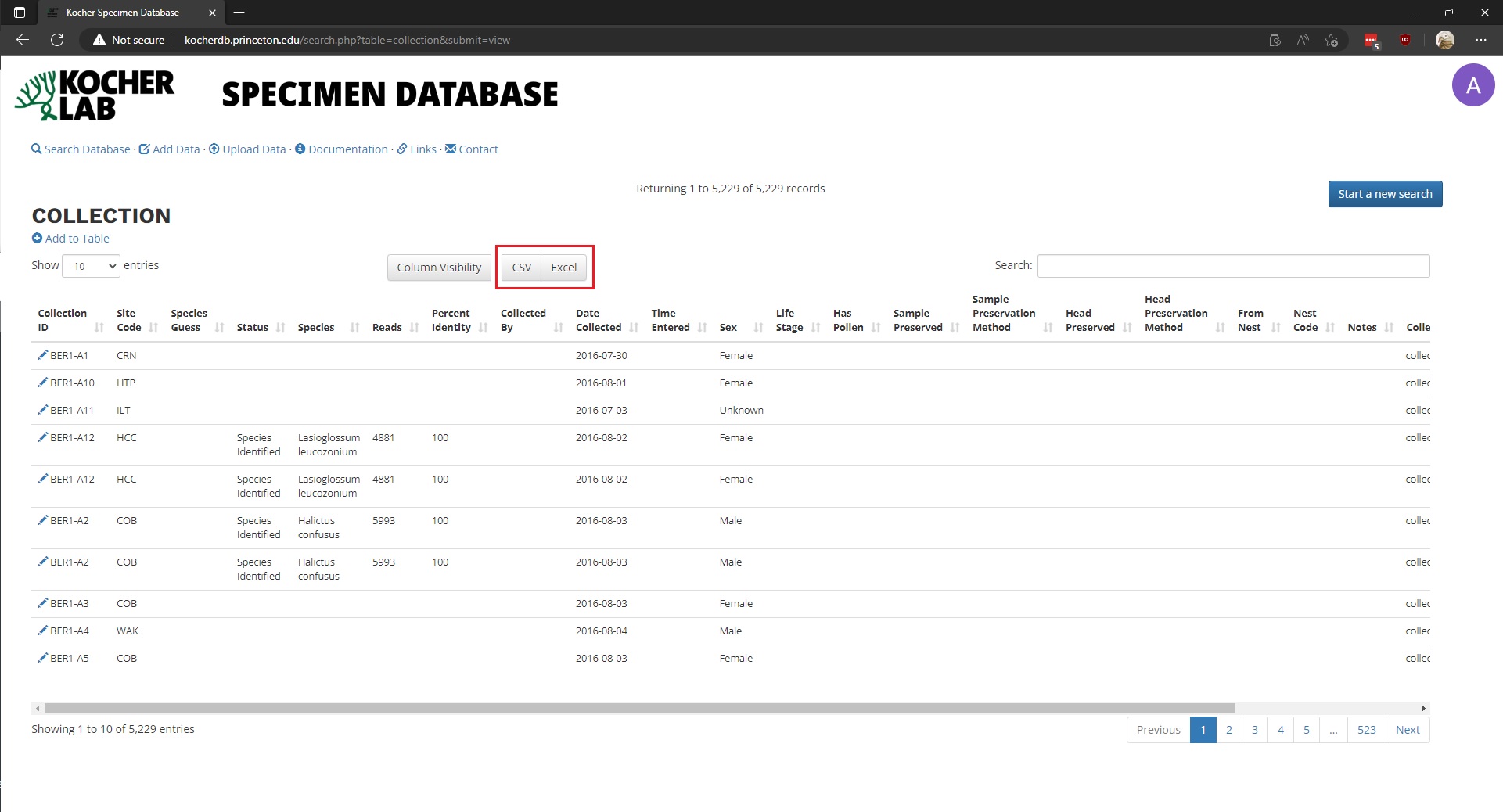
To begin searching the database, select Search Database link and select the table you wish to examine.
Once a table is selected, it will be displayed below. The displayed data may be downloaded by clicking either CSV or Excel.
Beenomes: Genome Browser and Sequence Database¶
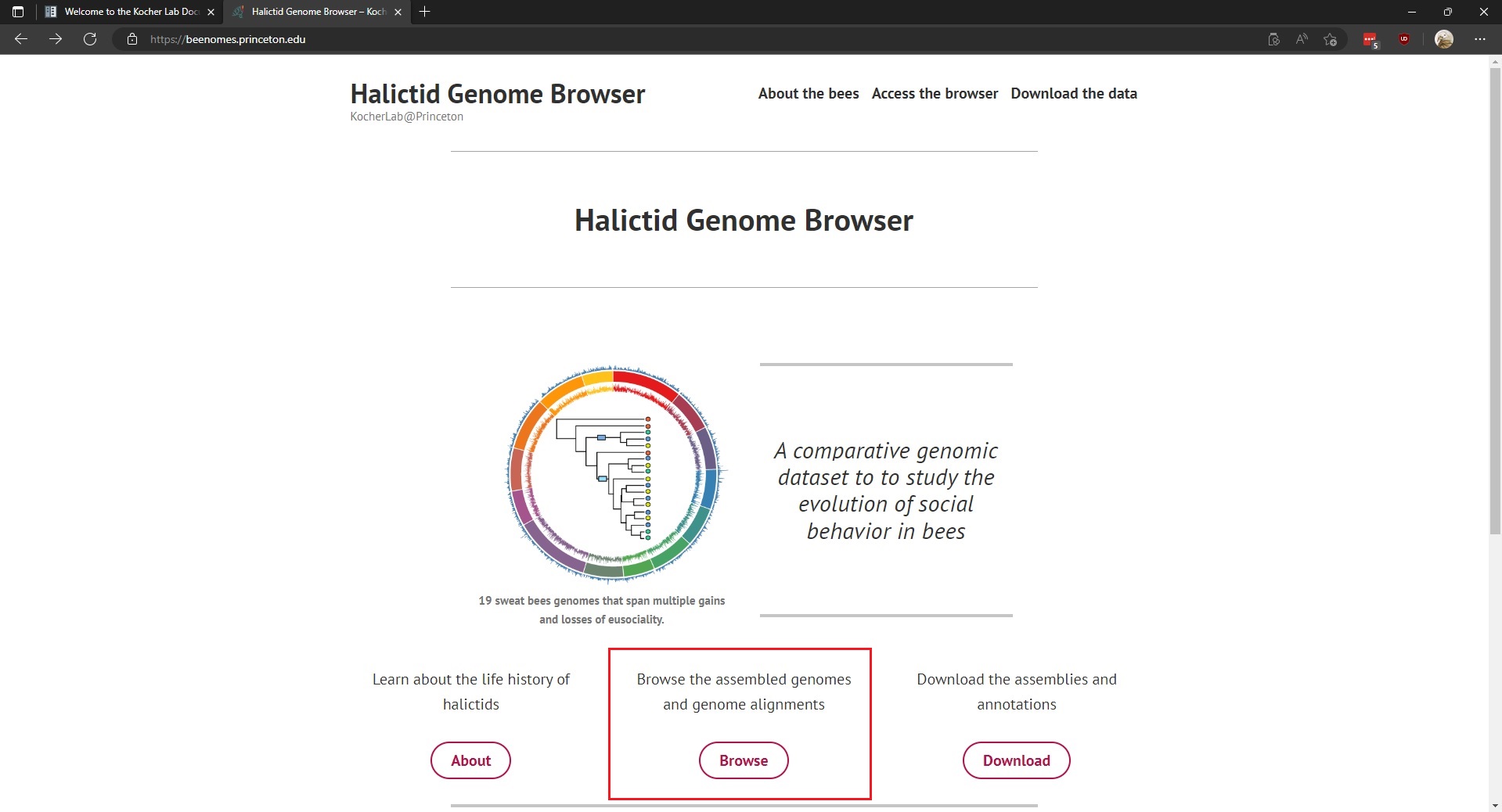
The Kocher lab genome browser and sequence database may be found at the Halictid Genome Browser. This page requires no special login and is available to the public. The homepage hosts links to access the genome browser and/or download the genomes (which are also available on Argo).
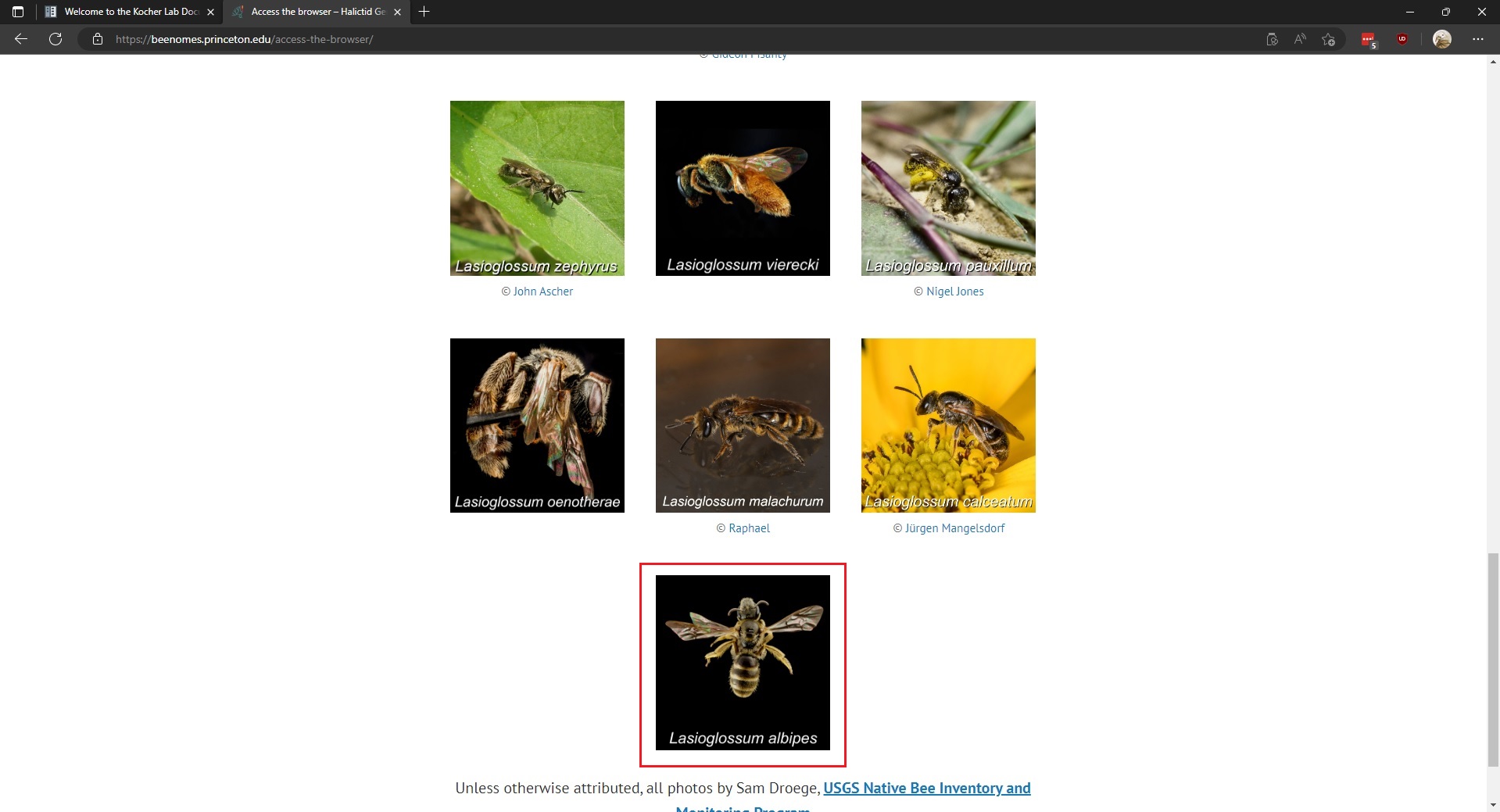
As an example, to access the genome browser for Lasioglossum albipes (LALB) you scroll until you reach the image of LALB, then click on the image.
The LALB genome browser (built using Jbrowse) will then be loaded. Like the UCSS genome browser, additional information may be displayed by selecting additional tracks.
It’s also possible to search the database for:
- Genes
- Ortholog Groups
- Chromosomal positions
If possible, the result will then be displayed.
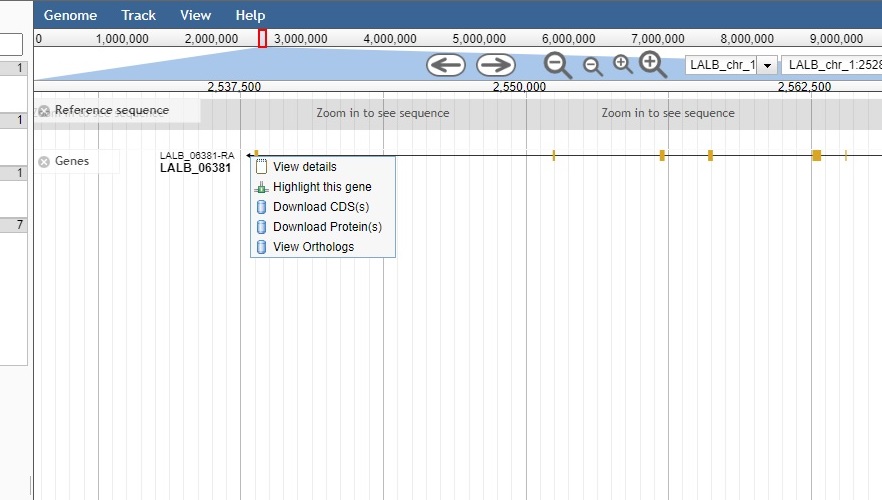
It’s also possible to simply zoom in to better display a gene of interest. To bring up additional information for a gene, one may:
- Left click to display relevant details
- Right click to display options, including links to the sequence database
The options listed are:
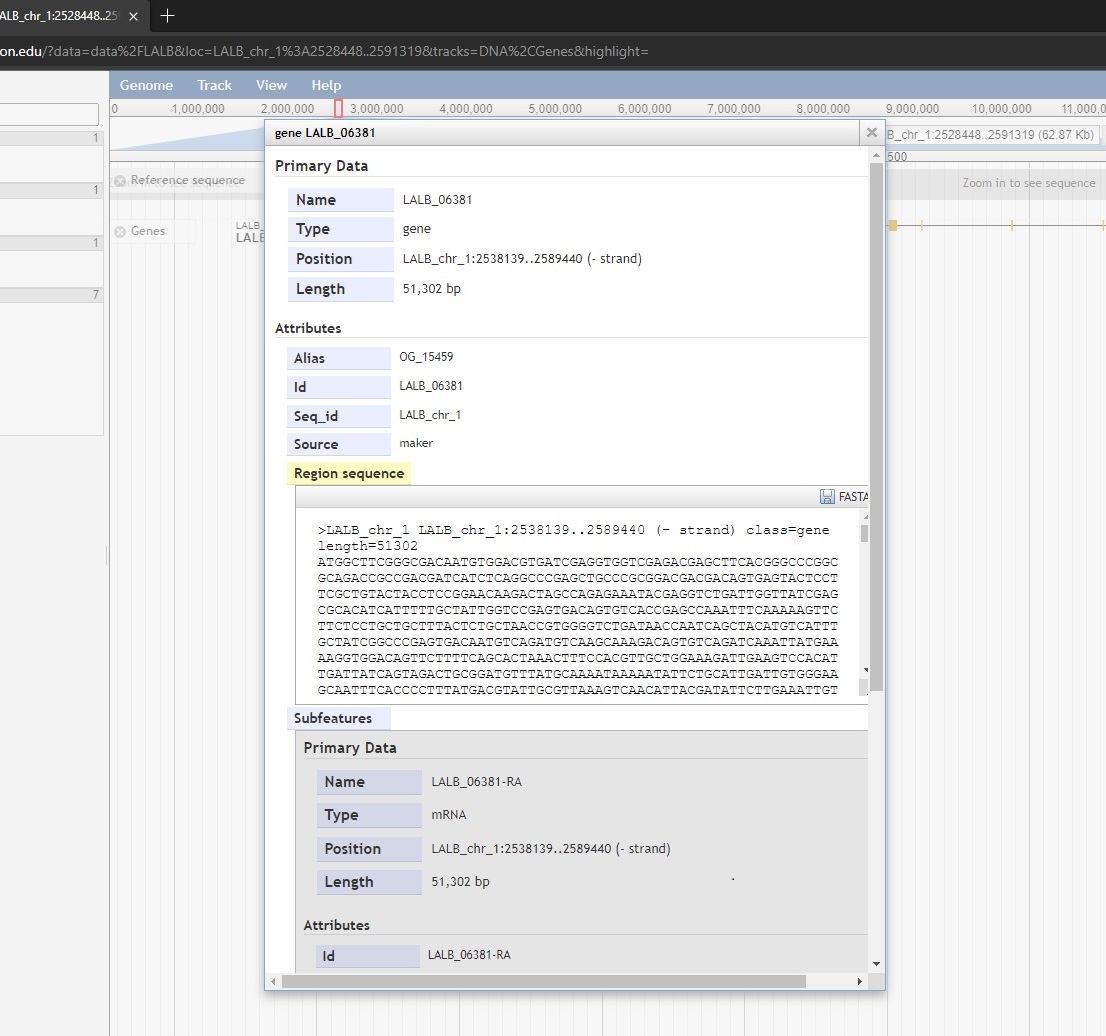
- View details – i.e. left clinking the gene
- Highlight the gene
- Download CDS(s) CoDing Sequence)
- Download Proteins(s)
- Download Orthologs
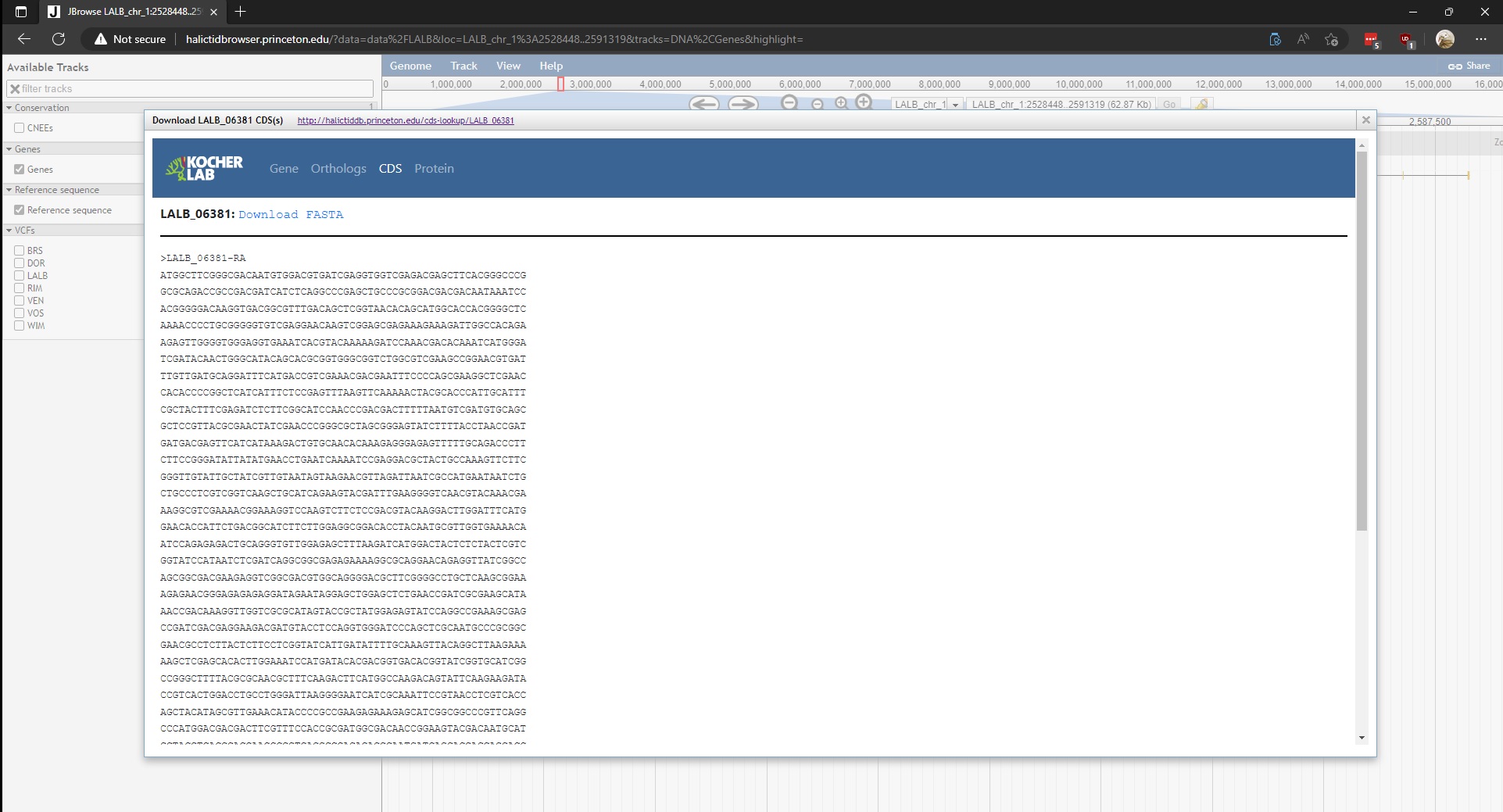
The three Download options open a new page on the Kocher lab sequence database. The sequence database was designed to store FASTA sequences (CDS and amino acid) and orthologs for each transcript. Selecting Download CDS(s) will open the following database entry for the gene in question.
At the top of the sequence database webpage there are links to:
- Orthologs – Site linking to the IDs of orthologs sequences (where possible).
- CDS – CDS sequence(s) of the of the gene in question. Also has an option to download FASTA file(s)
- Protein – Protein sequence(s) of the of the gene in question. Also has an option to download FASTA file(s)
- Gene (At present links to other information)
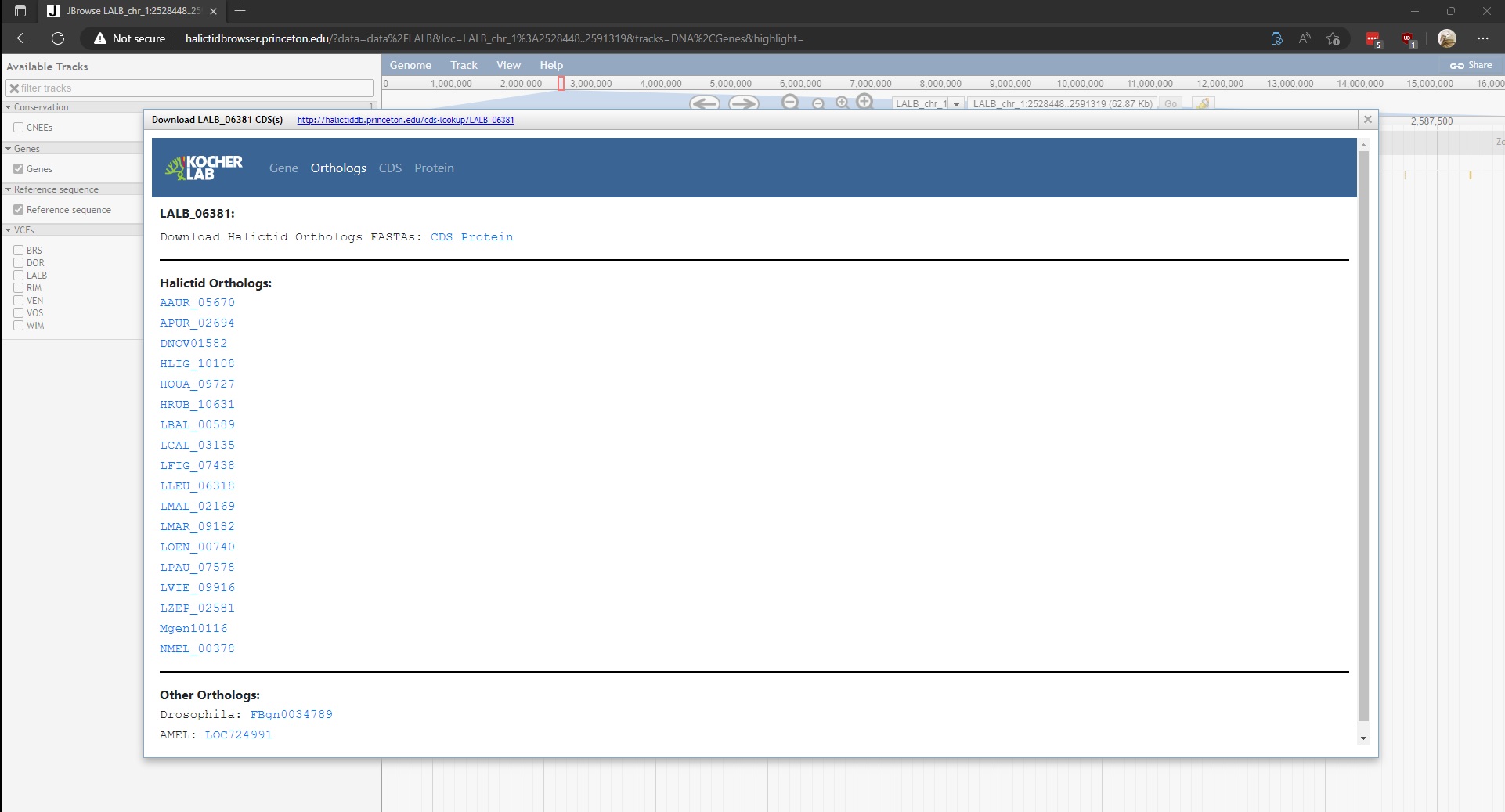
Selecting Orthologs will display the following page. Please note that Orthologs page displays both Halictid Orthologs and Other Orthologs (Drosophila and/or AMEL IDs). The page provides links to database entries for the Halictids and AMEL, and provides links to Flybase for Drosophila IDs. The page also provides links to download a FASTA file of the orthologs sequences.